Phylogenetic Networks Reimagined! Coming soon in Winter '25

Versatile Framework
Our platform offers a versatile framework and a robust set of data structures for developing customized phylogenetic network inference methods, enabling researchers to tailor solutions to their specific research needs.
Open Source Collaboration
PhyNetPy fosters open collaboration within the scientific community, allowing researchers to contribute and enhance the framework for mutual advancement in phylogenetic analysis.

Advanced Inference Methods
Benefit from our cutting-edge inference methods that analyze complex evolutionary scenarios and uncover intricate relationships in phylogenetic networks.
Committed to Performance
We never settle for good enough. For too long, researchers have been accepting of the fact that some network methods and anaylses can take upwards of many hours or even days to complete. With PhyNetPy, we have squeezed every ounce of performance out of our algorithms that we can.
MCMC-SEQ 2.0
Coming soon in release 1.0.0
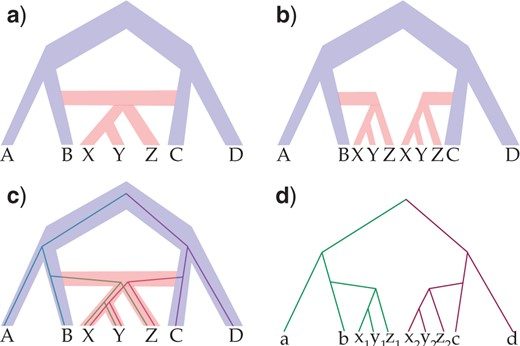
MP-Allop 2.0
Using a novel network search space move developed by Dr. Huw Ogilvie and Mark Kessler along with caching techniques, drastic runtime improvements on the scale of 100x the PhyloNet version have been recorded. Plus, support for autopolyploidy will be provided in an upcoming release.
InferNetworkMPL 2.0
Coming soon in release 1.0.0
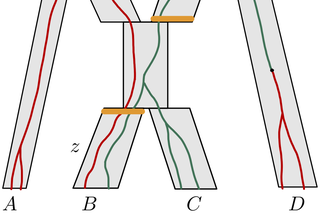
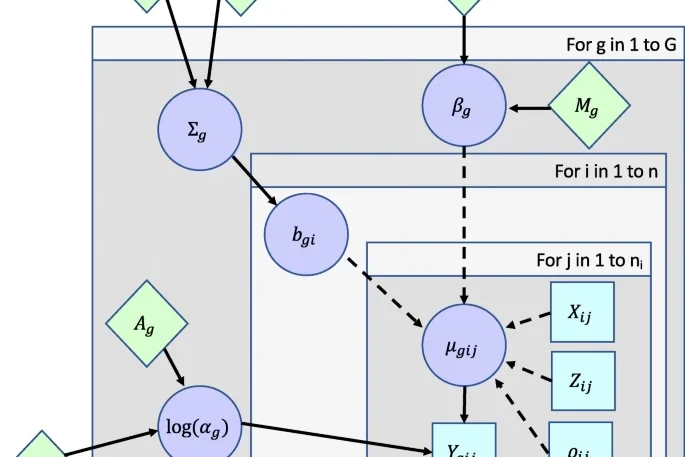
MCMC-BiMarkers 2.0
Built using a combination of a C# backend and Python frontend, this algorithm has been retooled in large part due to improvements made to the algorithm developed by Rabier et al. in the paper "On the inference of complex phylogenetic networks by Markov Chain Monte-Carlo"
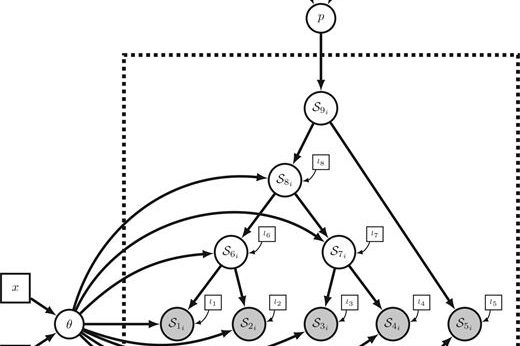
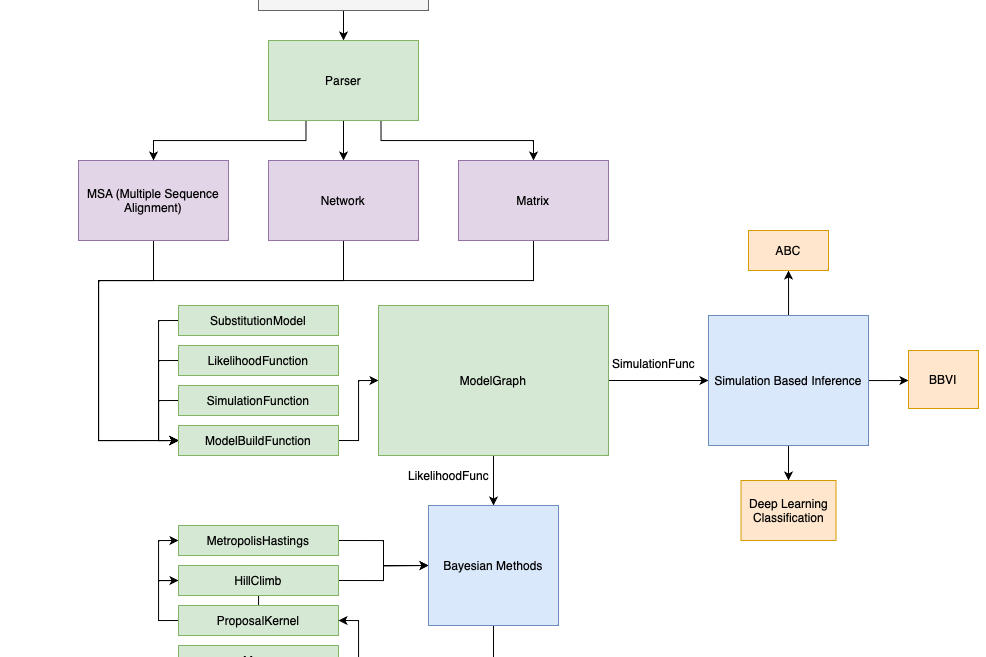
Probabilistic Graphical Modeling and Caching
The core of PhyNetPy is built around a probabilistic graphical modeling concept that allows for quick model prototyping, caching performance, and modularity that takes the pressure off the coder to make design decisions.
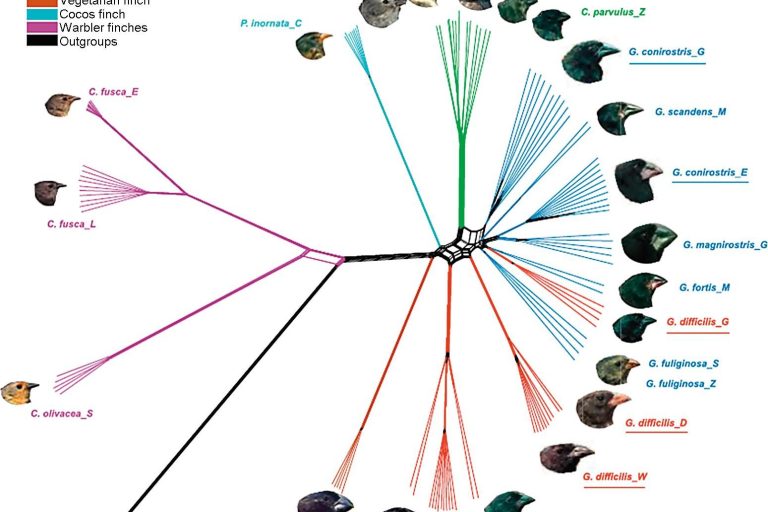
Diagnostic and Visualization Toolkit
Previous software packages for the field of phylogenetics lack integrated network tools for analyzing method results. We will also be bringing visualization functionality in a future release.
We need your consent to load the translations
We use a third-party service to translate the website content that may collect data about your activity. Please review the details in the privacy policy and accept the service to view the translations.


